JamieJQuinn Research Software Engineer
Analysing the prevalence of continuous integration in JOSS
JOSS is the Journal of Open Source Software and can be considered a collection of some excellent scientific codebases. Whether they’re exemplars of good coding practices is up for debate but I wanted to know roughly how many published papers (and associated codebases) were using specifically continuous integration of some form. Luckily I have some experience scraping web pages with python and the excellent webscraping package BeautifulSoup, so I cracked out the old tutorials and set to work.
Find the full source code in this Github repo.
Scraping the index
Webscraping is fundamentally very simple. The page is downloaded using the requests library:
def get_response(page_url):
"""Returns response to html request on page_url"""
response = requests.get(page_url)
return response
Then that’s loaded into BeautifulSoup with
soup = BeautifulSoup(response.content, 'lxml')
which produces a tree structure which can be searched with find and find_all and moved around using e.g. .link or .parent. In my case (as we’ll find out in the following paragraphs), I could extract all the paper URLs using
paper_entries = soup.find_all("entry")
paper_urls += [entry.link.get('href') for entry in paper_entries]
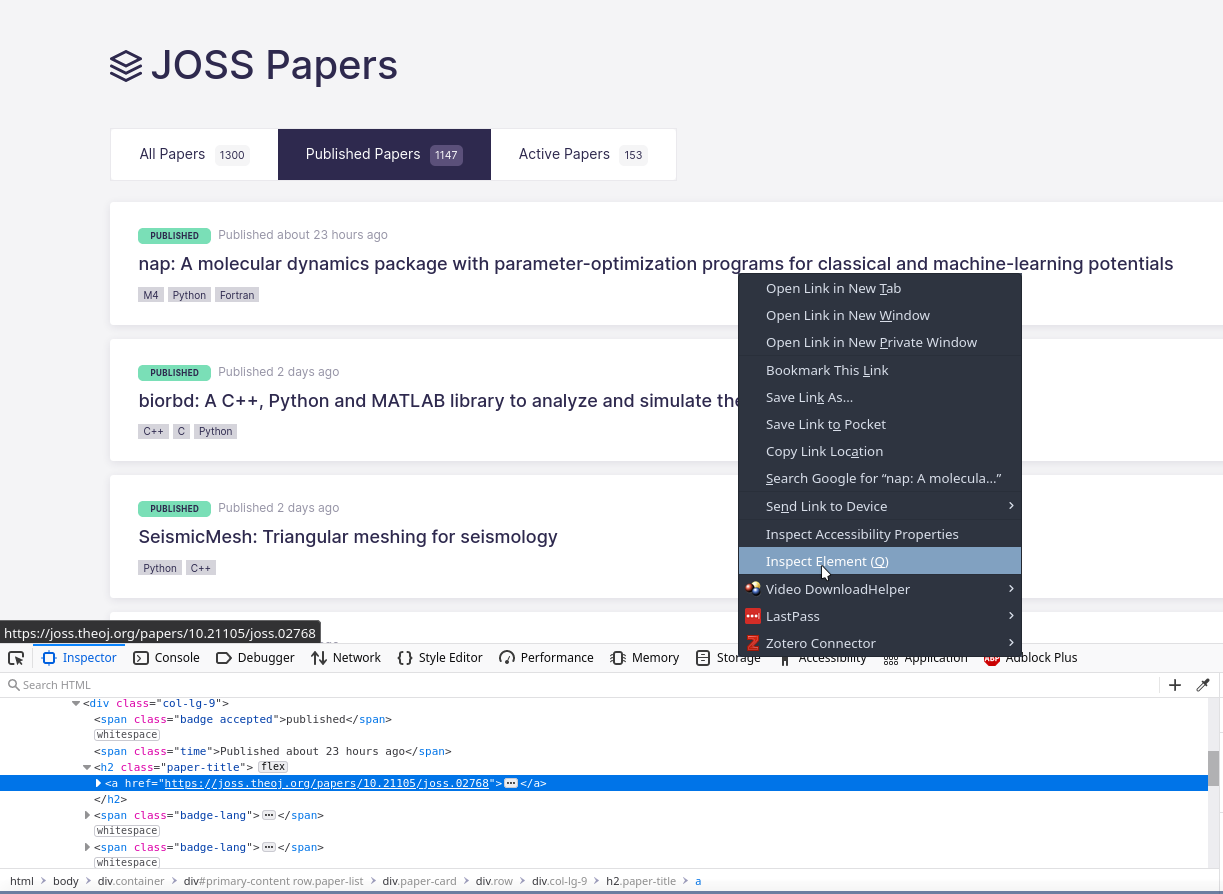
To find out how to parse a particular site, you have to get your hands dirty and start inspecting the HTML of the page you want to scrape. In my case, I wanted to know the URLs of each item in the index of JOSS so I went to the page in Firefox, right-clicked on the title of an item and clicked Inspect Element. This brings up a useful little HTML inspector tool which allows you to figure out how to identify different elements of the page (see screenshot below).
In my case I wanted the URL of the page associated with the paper (“nap” in the screenshot). That’s stored in the href of the <a> tag inside the <h2> title which is of the class ‘paper-title’. That relationship allowed me to construct a query for all matching <h2> tags, paper_entries=soup.find_all('h2', class_='paper-title'), and then extract the href from the <a> child. Putting that all together gave me a BeautifulSoup path which was completely incorrect.
Turns out the JOSS web server was recognising the request was coming from a non-browser and returned the atom feed instead. Nicely, BeautifulSoup can also parse those so after printing it out it was clear that each entry was kept in a <entry> tag and the link to the individual paper page stored in the <link> tag.
The URLs were then saved to a text file:
with open("ids.txt", 'w') as fp:
for item in paper_urls:
fp.write("%s\n" % item)
What I then realised was each URL was nearly identical barring a single ID at the very end. By stripping out the IDs using
cat paper_url_list.txt | awk -F. '{print $5}' > id_list.txt
I could reform the URL easily and use the ID as just that; a unique ID.
I should note that much of the above code was wrapped in a loop which iterated through the pages of the index (i.e. "https://joss.theoj.org/papers/published?page="+str(i)). See the Github repo linked at the top of the page for the full source code.
Scraping the metadata
Now I had a full index of all the paper IDs, I wanted to be able to scrape metadata from the associated URLs individually. One option was to load this list into python and loop over the list, scraping and saving the data for each paper. However, if it crashed for whatever reason, I didn’t want to lose progress and it can be a pain to parallelise tasks like this in python (not really true, there’s the Pool part of the multiprocessing library but it’s still more complex than I’d like). I realised I could use the tool make to solve both problems since it has inbuilt parallelisation (with the -j flag) and will automatically recognise when a file has been created (i.e. the output from the python script in this case) and will not run the same process again, that is it has some concept of state.
To fit into a make workflow I wrote a python script which takes one single paper URL, scrapes the page and spits out JSON-formatted metadata into a file so that it represented one single atomic operation. I then wrote a simple makefile which took in the list of IDs, formed the URLs and used the python script to output the associated metadata:
ID_FILE=data/id_list.txt
IDS=$(file < $(ID_FILE))
JSON_FILES=$(addprefix data/,$(addsuffix .json,$(IDS)))
all: $(JSON_FILES)
data/%.json:
python ./fetch_paper_details.py https://joss.theoj.org/papers/10.21105/joss.$* > $@
Running make -j4 on my 4-core laptop massively speeds up the scraping of over 1000 webpages. The python script loaded the URL, parsed it using BeautifulSoup and extracted the title from the meta div
meta = soup.find('div', class_='paper-meta')
title = meta.h1.string
the dates using the dateutil library
date_spans = meta.find_all('span', class_='small')
date_strings = [
parse(" ".join(span.string.split()[1:]))
for span in date_spans
]
extracted the tags associated with the languages used in the paper
lang_spans = meta.find_all('span', class_='badge-lang')
lang_tags = [span.string for span in lang_spans]
extracted the tags describing the field in which the paper sits from the sidebar of the page
sidebar = soup.find('div', class_='paper-sidebar')
tag_spans = sidebar.find_all('span', class_='badge-lang')
field_tags = [span.string for span in tag_spans]
and finally extracted the repository URL which just happens to be the first link which matches the search criteria
repo_url = soup.find('a', class_='paper-btn').get('href')
This last line is quite fragile. I wouldn’t necessarily depend on it to work if the page changed in future, but it works now and I only want the data once! All this was stored in a dict then saved as a JSON file
data = {
"title": title,
"submission_date": str(date_strings[0]),
"publish_date": str(date_strings[1]),
"lang_tags": lang_tags,
"field_tags": field_tags,
"repo_url": repo_url
}
Determining the CI platform
Now that I had a directory filled with JSON files each describing the metadata of one single paper, including the URL of the codebase repository I wondered how many papers actually stored their code in Github. Using cat *.json | sed 's/, /\n/g' | grep repo_url | awk -F'/' '{print $3}' | sort | uniq -c and some manual cleaning produced
17 bitbucket.org
1 c4science.ch
1 doi.org
1 framagit.org
1086 github.com
1 git.iws.uni-stuttgart.de
22 gitlab.com
1 gitlab.gwdg.de
3 gitlab.inria.fr
1 gricad-gitlab.univ-grenoble-alpes.fr
1 marcoalopez.github.io
1 mutabit.com
1 savannah.nongnu.org
1 sourceforge.net
3 ts-gitlab.iup.uni-heidelberg.de
1 www.idpoisson.fr
which strongly shows Github is the dominant platform for JOSS submissions and means I only have to write a scraper targetting one platform.
I looked at the source for a Github repository’s homepage and discovered a file in the repo appears in the webpage as a simple link with the title matching the filename in the repo, so I can search for, say, a Travis config file .travis.yml using
soup.find('a', title='.travis.yml')
After looking through around 30 of the most recent JOSS published papers, I could only find 3 continuous integration platforms in obvious use: Circle CI, Travis and Github Actions. These were signalled using
if soup.find('a', title='.circleci'):
# Is there a folder called .circleci?
return "circle-ci"
elif soup.find('span', string='.github/'):
# GH renders a github actions folder as a <span>
return "github-actions"
elif soup.find('a', title='.github'):
# GH renders a .github folder as an <a>
return "github-folder"
elif soup.find('a', title='.travis.yml'):
# Is there a file called .travis.yml?
return "travis"
else:
return False
with an additional option in case the repo wasn’t actually stored in Github. Note, where .github was stored in a <span> tag, that represented the situation where the .github folder only contained a workflows folder, indicating the project was using Github Actions. Where it was stored in a <a> tag instead, that represented the situation where other files were stored in the .github folder. For the sake of simplicity, I’m just counting those as also using Github Actions, even though that may not be true.
Again I used the make workflow to parallelise this whole process and the result was one file per paper (named [ID]_github.txt) storing the result of the above analysis.
Results
Using cat *_github.txt | sort | uniq -c gave a quick count of the number of papers using each platform and the results are stark:
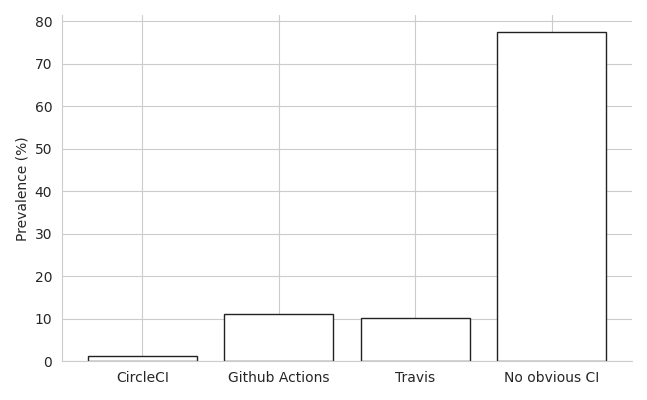
Or in raw numbers:
13 circle-ci
50 github-actions
72 github-folder // Could be github actions
110 travis
845 False // Couldn't find obivous CI
55 not-github
What we can draw from this is that the majority of projects do not use an obvious CI platform. Of course, some JOSS codebases are not stored in Github and some may be using webhooks or other tools to run alternative CI workflows like with Jenkins. Still, the sheer volume of codebases without any obvious CI setup is at least indicative of poor adoption of CI, even in codebases which have been peer-reviewed and published in an academic journal.
Thank you for reading this rather long and rambling recount of my recent obsession with continuous integration adoption levels. I hope you take away something from this, whether the use of make as a great tool for simple parallelisation, or BeautifulSoup as a fantastic webscraper, or some motivation to setup CI in your project before you become a statistic in my project. This entire analysis has many caveats and only rudimentary discovery of the CI platform attached to a codebase. If you have any ideas for improving this, please get in touch via twitter or by email.
PS. If you’re associated with JOSS, if there’s already an API then I’m sorry for thrashing your servers, but if there’s not… should there be?
Written on January 20th, 2021 by Jamie Quinn